Network Search Web UI
This document introduces the web interface of the INDRA Network Search Service
The network search interface with no input or results.
Source and Target
The source and target are the nodes between which to find a path and at least one of source and target is needed to do a search. If only one of source or target is provided, an open ended breadth first search is done instead of a path search. Note that the source and target are not affected by the choice of allowed namespaces (see below at Allowed Node Namespaces).
Autocompleting source/target inputs
Autocompletion of source/target based on prefix and entity identifier are made automatically as an input is typed or pasted into the text boxes. The suggestions are picked from the nodes in the graph and the text box will mark the entered text as correct if it matches an existing node in the graph.
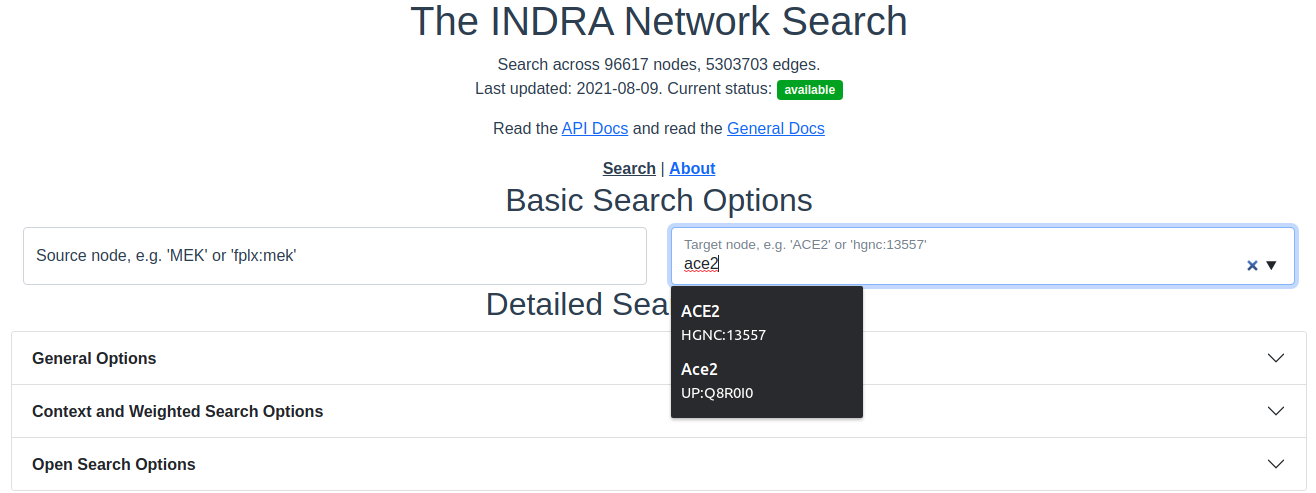
Autocompleting an entity. As an entity name is typed into the source or target text boxes, suggestions from the graph nodes are provided.
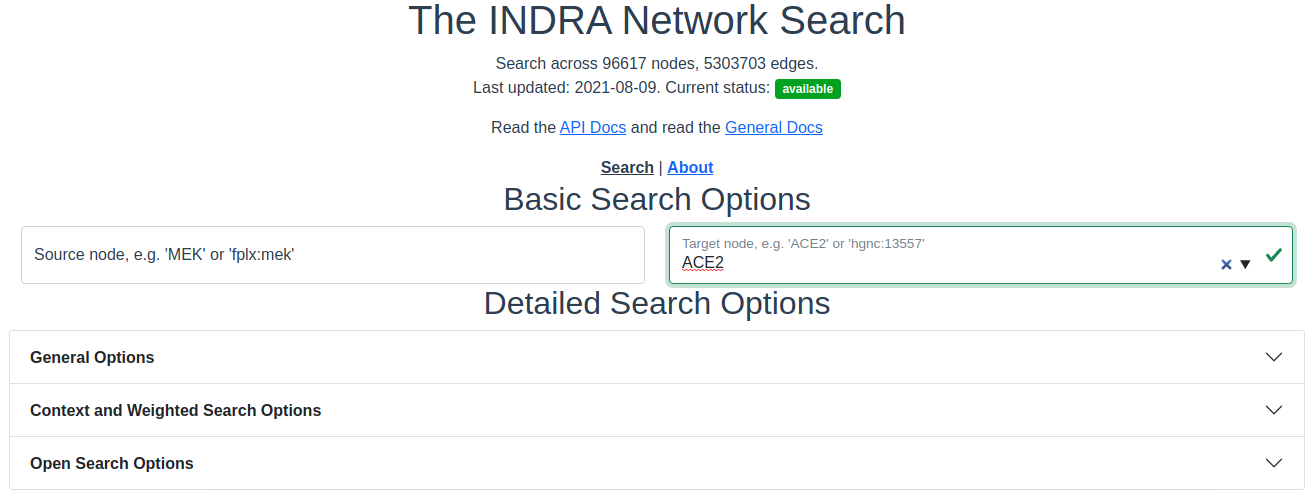
Verified entry. Entries are verified as they are being typed or pasted into the text box. When the entered entity is verified to exist in the graph, the text box border will switch to green and a checkmark will appear.
Detailed Search Options - General Options
The general detailed search options contain filters that apply to most searches, regardless of weighting or openness.
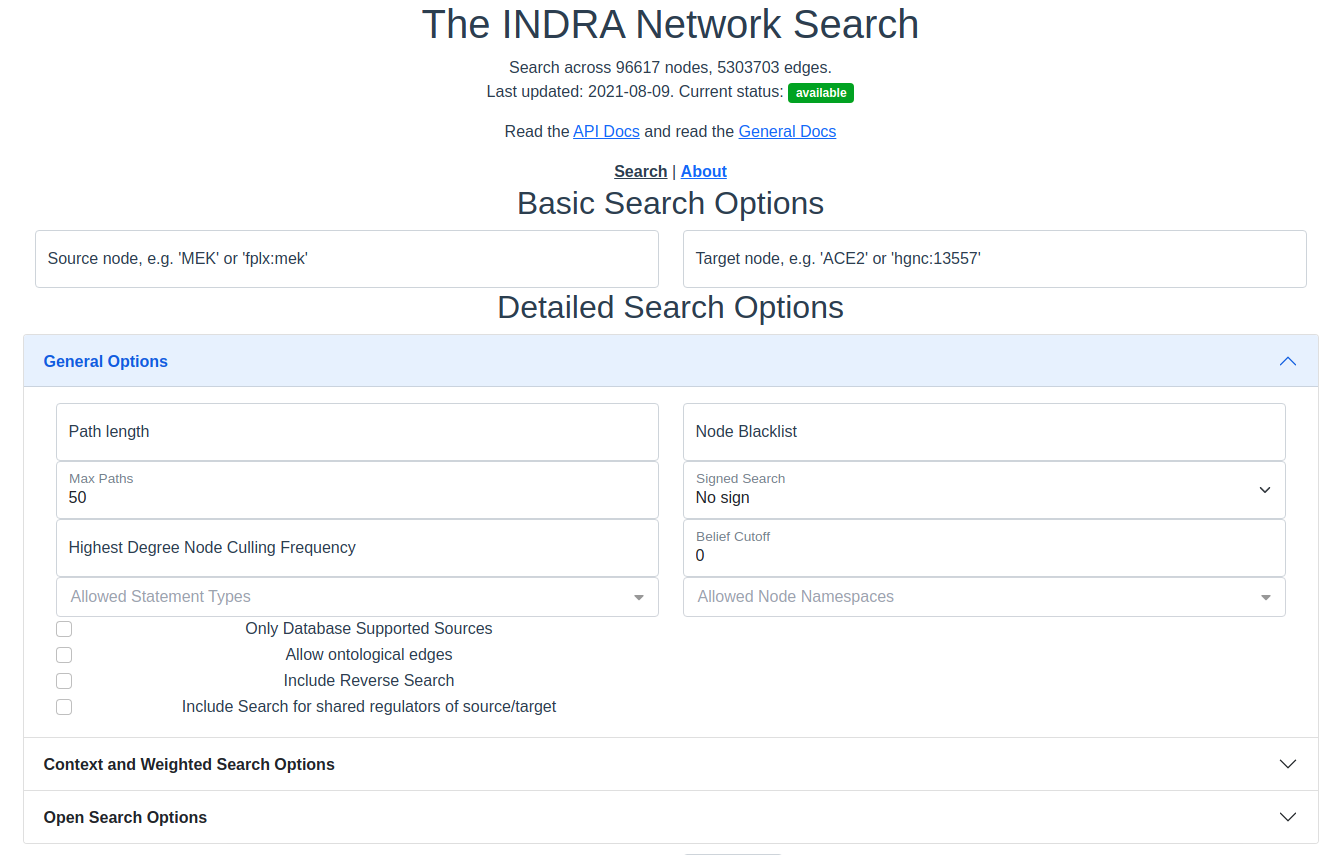
The search interface with the general options section expanded.
Path Length
Only paths of this many edges will be returned. Must be a positive integer.
Node Blacklist
Node names entered here are skipped in the path search. This is a good way to avoid nodes of extremely high degree that overwhelms the results and effectively blocks out results that include nodes of lower degree. See also Highest Degree Node Culling Frequency below.
Max Paths
The maximum number of results to return per category in the results. The default and the maximum allowed is 50 results. For unweighted searches this number rarely makes a perceivable difference in response time but for weighted searches keep this number low for a faster response time.
Signed Search
To perform a signed search, click on the drop down menu that says “No sign” and chose a sign. “+” means that the returned paths are upregulations, and “-” means that the returned paths are downregulations. For the purpose of signed search, only statements that imply a clear up- or downregulation are considered. Currently this mean IncreaseAmount and Activation for upregulation, and DecreaseAmount and Inhibition for downregulation.
Highest Degree Node Culling Frequency
Entering a positive integer here allows the path search to include the highest degree node for the first N returned paths, after which that node is added to the Node Blacklist. This is repeated for the second highest degree node for the following N paths, then for the third highest degree node and so forth. Limitations: This option is only applied to unweighted open search and source-target searches.
Belief Cutoff
Any statement with a belief score below this number will be excluded from the edge support. If all statements are excluded from the edge, all paths containing that edge become invalid and are skipped. It is set to zero by default to include all edges. Read more about belief scores in the belief module of INDRA.
Allowed Statement Types
This is a multiselect dropdown which contains multiple statement types to allow in the results. If an edge of a path does not contain any of the selected statement types, the whole path will be skipped from the result. Read more about statement types in the statements module of INDRA.
Allowed Node Namespaces
The namespaces included here are the ones that are allowed on any node visited in the path search. The namespace of the source and target are excluded from this restriction. A namespace in INDRA is the prefix or name of the type of identifier used to uniquely identify an entity from a specific knowledge source. For example, a chemical can be identified using a CHEBI identifier and would then be identified in the CHEBI namespace.
Checkboxes
The following options are available as checkboxes:
Only Database Supported Sources: Check this box to enforce that all edges must be supported by at least one statement sourced from curated databases like PathwayCommons and Signor
Allow Ontological Edges: Check this box to allow directed edges that go from an entity to its ontological parent, e.g. from the NFKB1 sub-unit to the NFkappaB complex.
Include Reverse Search: Check this box to also search for paths with source and target swapped. With this option, the reverse search from target to source is done as well as the original search from source to target. If the timeout is reached (see below) before the reverse search can start, the reverse search will not return any paths. If the timeout is reached during the reverse search, fewer paths than for the original search will be returned.
Include Search for Shared Regulators of Source/Target: Check this box to include a search for common upstream nodes one edge away from both source and target. This option is only available when both source and target specified.
Detailed Search Options - Context and Weighted Search Options
This section of the search options allows control over how to prioritize or weight edges in paths differently. During weighted search, the cost along every path encountered is calculated as the sum of the edge weights along the path. The paths are returned in ascending order of cost.
The different ways of weighting the search are available in the dropdown menu “Weighted Search”. Note: A weighted search is costly and usually takes longer than an unweighted search. It is common that a very heavy weighted search times out, especially for a signed weighted search, even with the highest allowed timeout of 120 seconds.
The weighted search uses a slightly modified version of the Djikstra weighted search employed in Networkx.
The code implemented for the weighted search is available on github in the functions shortest_simple_paths() and open_dijkstra_search() for closed and open paths, respectively.
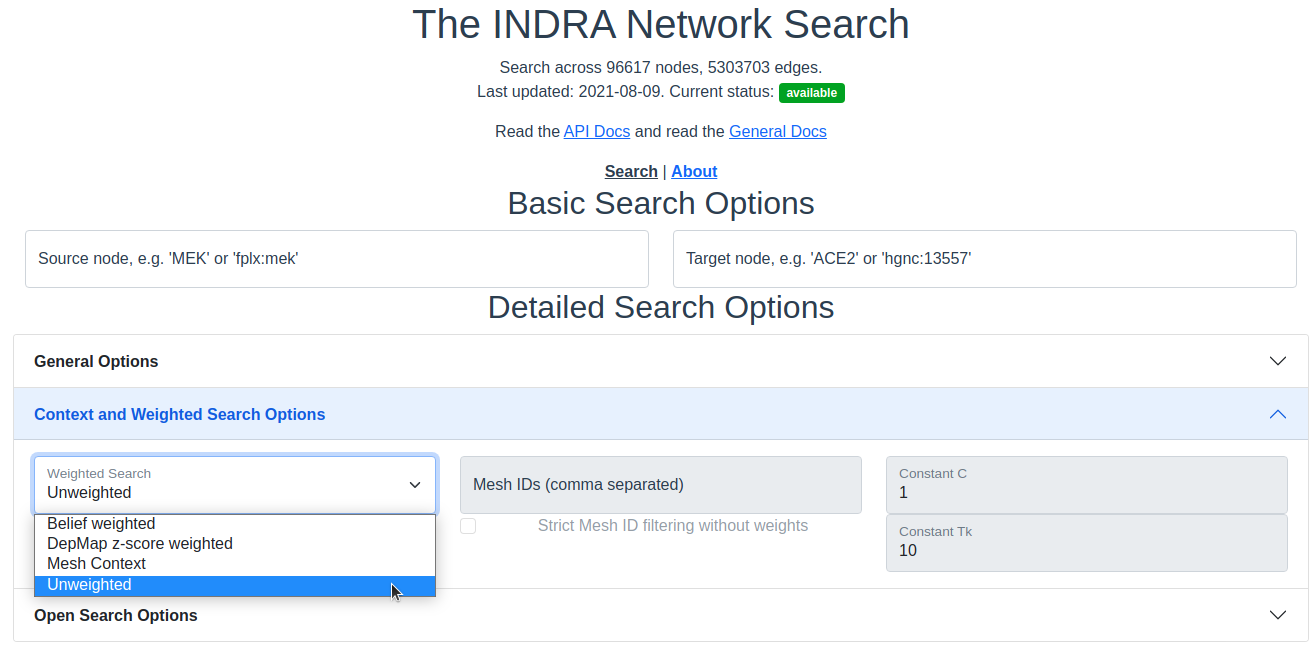
The search interface with the Context and Weighted search options section expanded.
Unweighted
This is the default option and imposes no weight on the edges and is equivalent to all edges having a unit weight.
Belief Weighted
The belief weight of an edge is calculated as the negative log of the aggregated belief scores of all the statements supporting edge \(e\):
where \(b_i\) is the belief score of supporting statement \(i\) of edge \(e\). Since the belief score is limited to the interval \([0, 1]\), it can interpreted as a probability and the above weight can therefore be seen as the log of the complement to the probability that every supporting statement is incorrect.
DepMap z-score weighted
The z-score edge weight is focused around prioritizing edges between human genes that have been targeted in knockout screens performed at the Broad Institute’s Dependency Map project. Stouffer’s method is used to get z-scores from the p-values of each correlation. The calculation is done on the log of the p-values for increased precision:
Here, \(\mathbf{R}\) is the correlation matrix of the gene expression data, \(\log (\mathbf{p})\) is the matrix containing the log of the p-values, and \(\text{logcdf}_{\text{beta}}\) is the log of the cumulative distribution function of the beta distribution. To get the z-score, we use the inverse of the log of the normal distribution’s CDF and then recover the sign from the original correlations:
with \(f\) being the inverse of the log of the normal distribution’s CDF, \(\left| \mathbf{z_e} \right|\) being the matrix of z-score magnitides, \(\mathbf{z_e}\) being the matrix of z-scores with sign and \(\text{sign}()\) mapping values to their signs.
The edge weight, assuming both nodes are human genes, is calculated by normalizing the difference between the z-score associated with a self-correlation and the strength of the z-score between the two nodes of the edge. In the case that one or both of the nodes of the edge are non-gene entities, the z-score weight is set to 1:
where \(z_0\) is the z-score associated with self correlation and \(z_e\) is the z-score of the edge.
Mesh Context
The context based search allows a search to prioritize or only include connections that are relevant to the provided context. The context is given as MeSH terms.
MeSH IDs: Enter the MeSH IDs, separated by comma, that should be used in the search.
Strict Filtering on MeSH IDs: Check this box to only allow edges with associated with the provided MeSH IDs. If left unchecked, the search is weighted.
Constants \(C\) and \(T_k\): These two constant allow for changing the importance of the context in a weighted context based search. For any edge \(e\), the weight \(w_e\) of the edge in the context based search is calculated in the following way:
Here, refcount is the number of references with the associated MeSH ID(s) that are supporting edge \(e\) and total is the total number of references supporting edge \(e\).
Detailed Search Options - Open Search Options
Options under the Open Search Options are only applied during open searches, i.e. when either of source or target is provided.
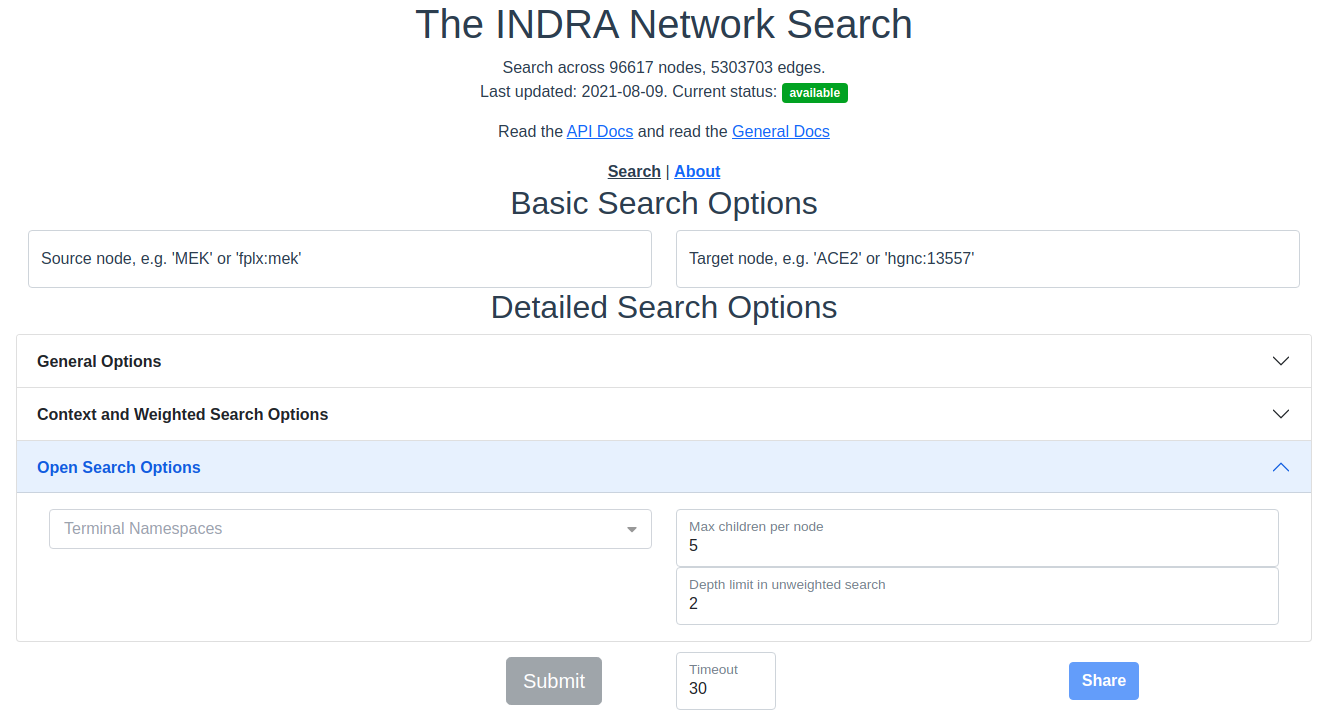
The search interface with the Open search options section expanded.
Terminal Namespaces
Namespaces selected here restrict the search to only return paths that end (open search from source) or start (open search from target) on the given namespaces and then not consider these nodes further. For example: if namespace A is selected, then a downstream path might look like this: X->Y->A, but not like this: X->Y->A->Z, where X, Y, Z are all namespaces other than A.
Max Children Per Node (Unweighted Search)
The integer provided here gives a maximum for the number of children to continue to open search from. For example: if N is set here, the first N nodes selected from the starting node are then considered for the next layer in the breadth first search. This option is only available for unweighted searches.
Depth Limit (Unweighted Search)
This option limits how deep, i.e. how many edges, the returned paths are allowed to be/have. This option is only available for unweighted searches.
Timeout
Setting a timeout allows to set a larger (or smaller) timeout than the default 30 seconds timeout. The time since the path search was started is checked before each iteration of data assembly for a returned path during the search. If the time passed is larger than the allowed timeout, the search is stopped. The timeout provided has to be a decimal number smaller than or equal to 120 seconds.
Result Categories
Note: If there are no results for a specific category, that section will be hidden.
Common Parents
This section shows the result of a search for common ontological parents of source and target. For example with GP1BA and GP1BB as source and target, respectively, the Platelet membrane glycoprotein complex shows up as a shared ontological parent.
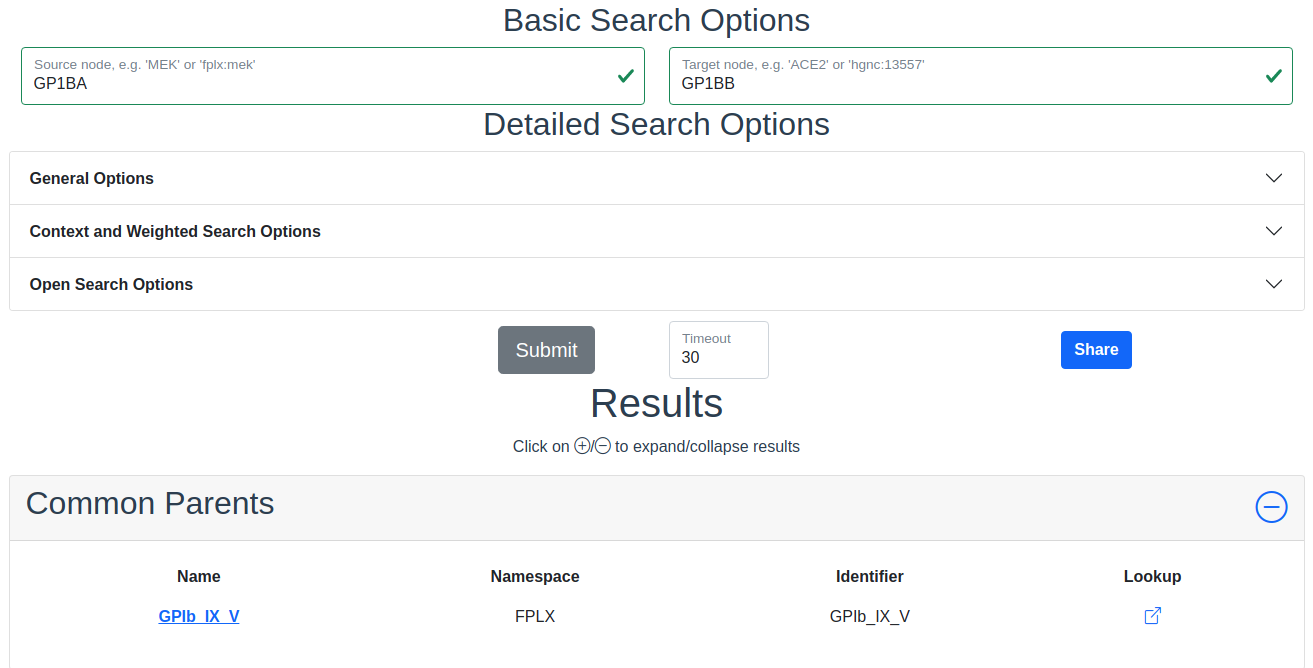
The result of a search with GP1BA and GP1BB as source and target, respectively, for Common Parents showing the Platelet membrane glycoprotein complex as their shared protein complex.
Path Results
This section show path results per path length, i.e. all paths with the same number of edges share a specific subsection. The division of paths per subsection is done regardless if the path search is weighted or not.
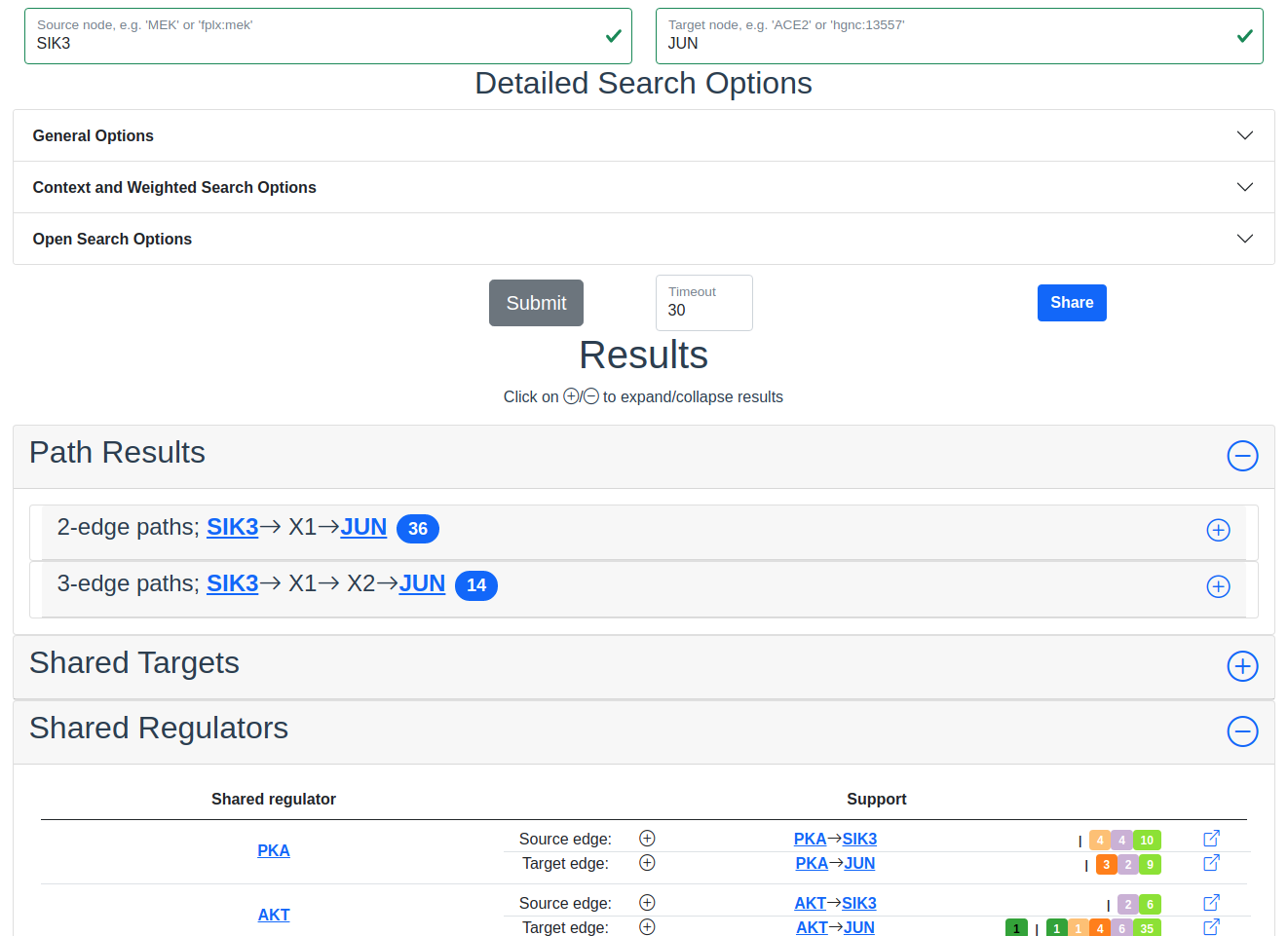
Search results with SIK3 as source and JUN as target with the Shared Regulator section expanded.
Detailed Results
For each result section, excluding Common Parents, there are two levels of detail. Results for Common Parents only have one level of results: name, namespace, identifier and entity lookup. The first level shows path (for Path Results), target (for Shared Targets) or regulator (for Shared Regulators) together with weight (if the search is weighted) the edge, source counts and a link to the INDRA DB for that specific edge.
The second level of results is collapsed by default. To expand it, the circled “+” (\(\oplus\)) need to be clicked. Once expanded, source counts and a link to more specific information in the INDRA DB per statement type are shown.
As the network search results can be filtered in more detail than what is possible using the INDRA DB, the statements shown in the DB can sometimes be a superset of the statements shown in the second level of the results.
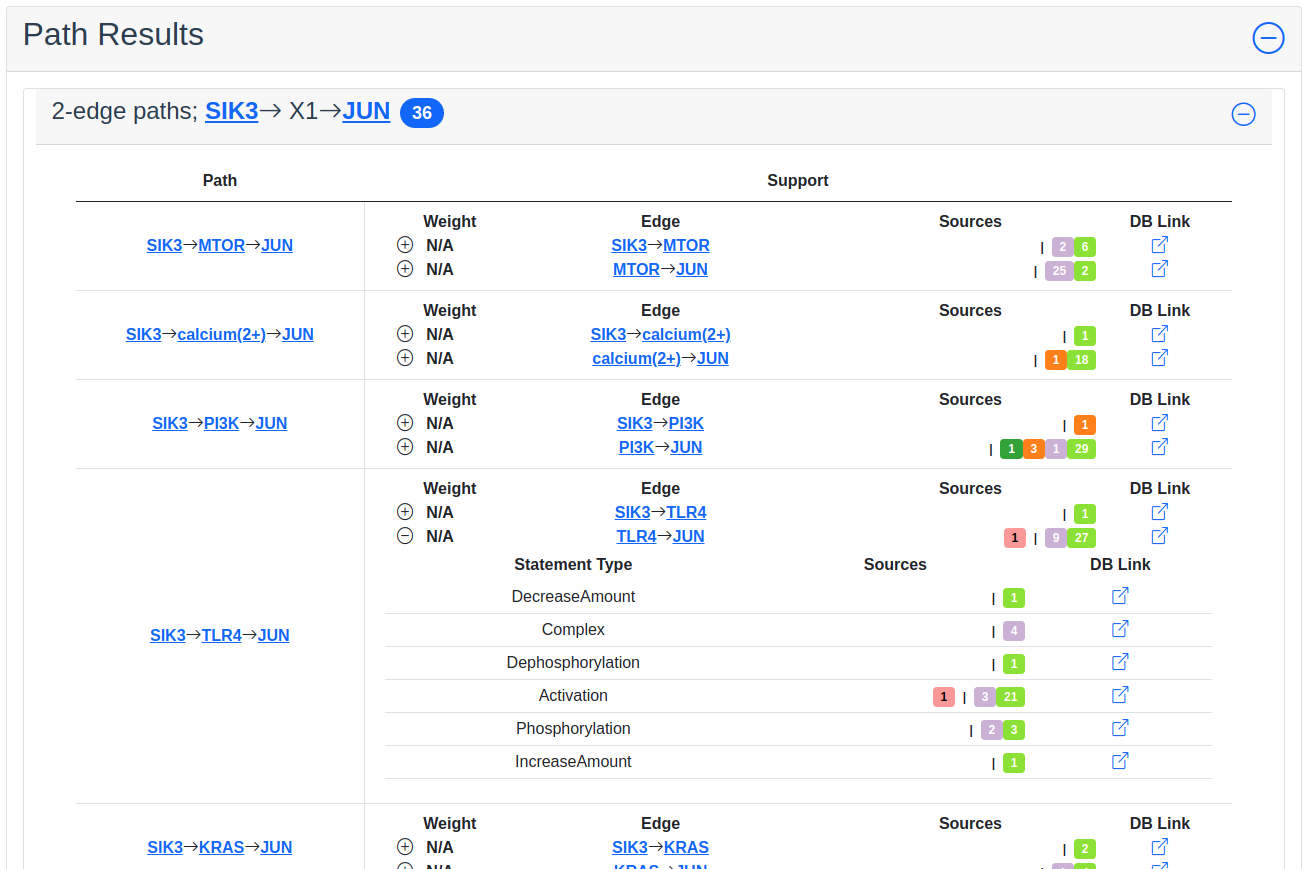
Path search results with one of the edges of a path expanded to more detail.
The Graphs Used
The two graphs used for the network search are assembled from a full snapshot of the INDRA DataBase that is updated regularly. Any statement that includes two or three agents are assembled into the support for the edges for the graphs, with one edge containing one or more statements. The two types of graphs used are:
Unsigned directed graph
Signed node directed graph
The edges in the signed graph only contain statements that have clear up- or downreguations associated with them, which currently are IncreaseAmount and Activation for upregulation, and DecreaseAmount and Inhibition for downregulation.
The code assembling the graphs can be found in net_functions.py in the function sif_dump_df_to_digraph() in the depmap_analysis repository.